alternative splicing
A cell typically splices a single transcript in multiple ways to generate an assortment of proteins. Alternatively spliced exons tend to lie between those segments of a gene that encode the functional units, or domains, of a protein (introns). An average mammalian gene possesses eight or nine exons—since most human genes undergoing some form of alternative splicing, virtually all of these exons are candidates for elaborate control.
Alternative splicing depends upon a splice site and nearby enhancer and repressor sequences—short segments of RNA that couple with regulatory proteins. It has been estimated that the splicing of a single exon may be promoted by at least three to seven enhancer sequences. As a result of alternate splicing, mutations that alter a splice site or a nearby regulatory sequence can have subtle effects by shifting the ratio of the resulting proteins without entirely eliminating any form.
Alternative splicing can allow one gene to generate different proteins in different tissues. Many highly specialized brain proteins arise from differential splicing of genes that are also expressed in other tissues. Cells can even modify splicing in response to changing conditions, and not only can alternative splicing tweak the structure of a single protein, but it may also be a means of regulating entire pathways. alternative splicing - click on fig 1 for animation : life cycle of an mRNA ~ click on Quicktime Q :
HHMI Feature Article on Alternative Splicing : Artist's conception of AS
Controlling the Synapse — 49 Proteins at a Time : The Alternative Splicing Website : Alternative Splicing DB (ASDB) : DNA-RNA-ProteinNational Center for Biotechnology Information

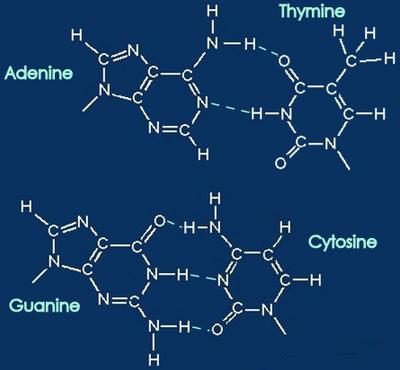 Above -
Above -